Thesis wordcloud
For one of my Covid-19 lockdown projects I wanted to create a wordcloud of my PhD thesis and as the subject of my thesis was neuroscience, I thought it would be fun to make it in the shape of a brain.
Environment setup & installation of packages
I use the terminal on MacOS and run Python through Anaconda but you should be able to run these commands with minimal adjustments on other systems.
Run the command below to initialise a new environment called wordcloud, install python, the wordcloud package and its dependencies.
conda create -n wordcloud python=3.7 pip wordcloud
conda activate wordcloud
pip install nltk pdfminer
Loading packages
import sys
sys.modules[__name__].__dict__.clear()
# Loading libraries
import numpy as np
import re # regular expression
from PIL import Image # [Python Imaging Library](https://pypi.org/project/PIL/)
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
from io import StringIO
import unicodedata
from nltk.corpus import stopwords
from wordcloud import WordCloud, ImageColorGenerator
# import packages from pdfminer
from pdfminer.converter ``import TextConverter
from pdfminer.layout import LAParams
from pdfminer.pdfdocument import PDFDocument
from pdfminer.pdfinterp import PDFResourceManager, PDFPageInterpreter
from pdfminer.pdfpage import PDFPage
from pdfminer.pdfparser import PDFParser
Importing the pdf
I used the package pdfminer to read my my PhD thesis into python. I limited the number of pages to import in order to exclude the pages with references.
page_numbers = (0, 135) # get pages up to references
# import pdf as text
output_string = StringIO()
with open('data/thesis_JochemVanKempen.pdf', 'rb') as in_file:
parser = PDFParser(in_file)
doc = PDFDocument(parser)
rsrcmgr = PDFResourceManager()
device = TextConverter(rsrcmgr, output_string, laparams=LAParams())
interpreter = PDFPageInterpreter(rsrcmgr, device)
for page_number, page in enumerate(PDFPage.create_pages(doc)):
if (page_number>=page_numbers[0]) and (page_number<=page_numbers[1]):
interpreter.process_page(page)
# print(output_string.getvalue())
new_string = str(output_string.getvalue())
Cleaning up the text
Next, I cleaned up the text by removing brackets, special characters and numbers. Finally, I made the text lowercase.
# Remove brackets
new_string = re.sub('[\(\[].*?[\)\]]', ' ', new_string)
# Remove line breaks
new_string = re.sub('\n', '', new_string)
# Remove page breaks
new_string = re.sub('\x0c', '', new_string)
# Remove accented characters and normalise using the unicodedata library
new_string = unicodedata.normalize('NFKD', new_string).encode('ascii', 'ignore').decode('utf-8', 'ignore')
# make lower case
new_string = new_string.lower()
Define a few acronyms, or make them upper case
Some acronyms I reverted back to upper case, others I defined.
# convert specific acronyms back to upper case
new_string = new_string.replace(' cpp ', ' CPP ')
new_string = new_string.replace(' eeg ', ' EEG ')
new_string = new_string.replace(' fef ', ' FEF ')
new_string = new_string.replace(' hmm ', ' HMM ')
new_string = new_string.replace('hz', 'Hz')
new_string = new_string.replace(' itpc ', ' ITPC ')
new_string = new_string.replace(' lc ', ' LC ')
new_string = new_string.replace(' lfpb ', ' LFPb ')
new_string = new_string.replace(' n2c ', ' N2c ')
new_string = new_string.replace(' rf ', ' RF ')
new_string = new_string.replace(' rt ', ' RT ')
new_string = new_string.replace(' v1 ', ' V1 ')
new_string = new_string.replace(' v4 ', ' V4 ')
# replace some acronyms with words
new_string = new_string.replace(' ach ', ' acetylcholine ')
new_string = new_string.replace(' da ', ' dopamine ')
new_string = new_string.replace(' na ', ' noradrenaline ')
Exluding words
Using the natural language toolkit package nltk, I obtained a list of stopwords to exclude from the wordcloud. I extended this list with some words specific to scientific referencing (for instance ‘doi’ and ‘et al’) as well as words specific to my thesis (for instance specific author names that I cited frequently)
from nltk.corpus import stopwords
stopword_list = stopwords.words('english')
stopword_list.extend(['et', 'al', 'doi',
'figure', 'chapter',
'and','well','thus','first','also',
'nat','neurosci','jneurosci','sci','science',
'one','two','although',
'thiele','aston','jones','cohen','arnsten','bellgrove','connell',
'non','within',
'single','additionally', 'p','ms','u', 'd1r', 'b', 'pre'])
Making/importing the mask
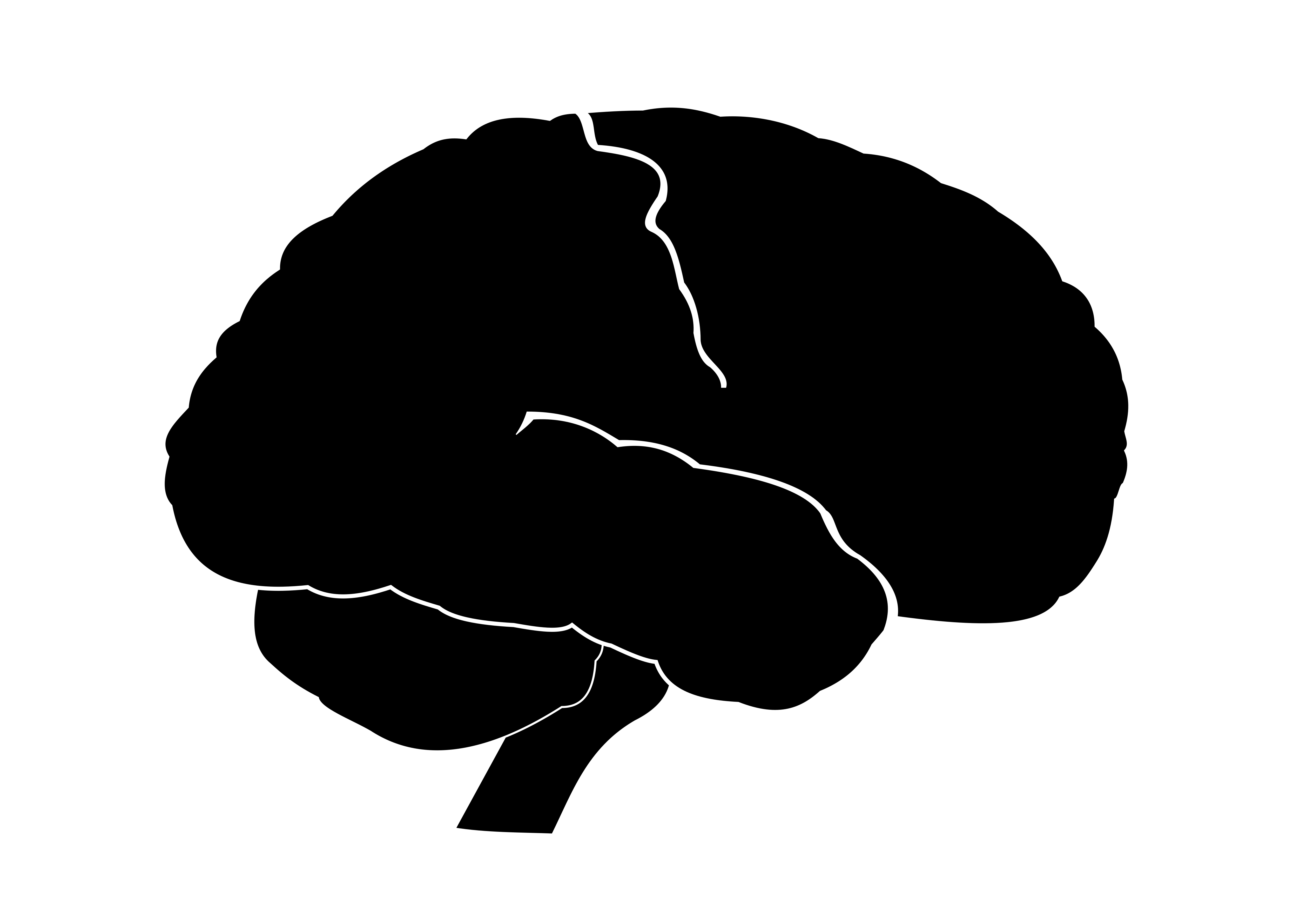
I created a mask for the wordcloud in the shape of a brain using inkscape by adapting this image. The WordCloud package uses all black values of the mask to plot words (RGB = [0 0 0]). I added a few white lines to mark the cerebellum and the temporal lobe so that these boundaries will still be visible in the wordcloud.
Importing the mask into python and plotting it:
mask_brain = np.array(Image.open("Img/brain_blackw.png"))
plt.imshow(mask_brain)
plt.axis("off")
plt.show()
Making the wordcloud
Making the wordcloud is fairly straightforward using the WordCloud package.
I chose a black background and to display the 200 most used words.
Make sure to set mask = mask_brain.
wc = WordCloud(stopwords = stopword_list,
background_color = "black",
mask = mask_brain,
collocations = False,
max_words=200, width = 1600, height = 900,
random_state=1).generate(new_string)
print(*wc.words_, sep = "\n")
Set the colour scheme
To change the colour scheme, I wanted to only use part of the bone colormap, from 0.5 to 1. For this, I used the function truncate_colormap that I found here.
# truncate colormap
def truncate_colormap(cmap, minval=0.0, maxval=1.0, n=-1):
if n == -1:
n = cmap.N
new_cmap = mcolors.LinearSegmentedColormap.from_list(
'trunc({name},{a:.2f},{b:.2f})'.format(name=cmap.name, a=minval, b=maxval),
cmap(np.linspace(minval, maxval, n)))
return new_cmap
minColor = 0.5
maxColor = 1.00
cmap2use = "bone"
cmap_t = truncate_colormap(plt.get_cmap(cmap2use), minColor, maxColor)
The final result
Finally, I plotted the wordcloud with the truncated colormap.
plt.imshow( wc.recolor(colormap = cmap_t, random_state = 1), alpha = 1 , interpolation='bilinear')
plt.axis("off")

Code availability
The code I used to create this wordcloud is available on my gitlab page
Other tutorials
I used the following tutorials which you might find useful:
